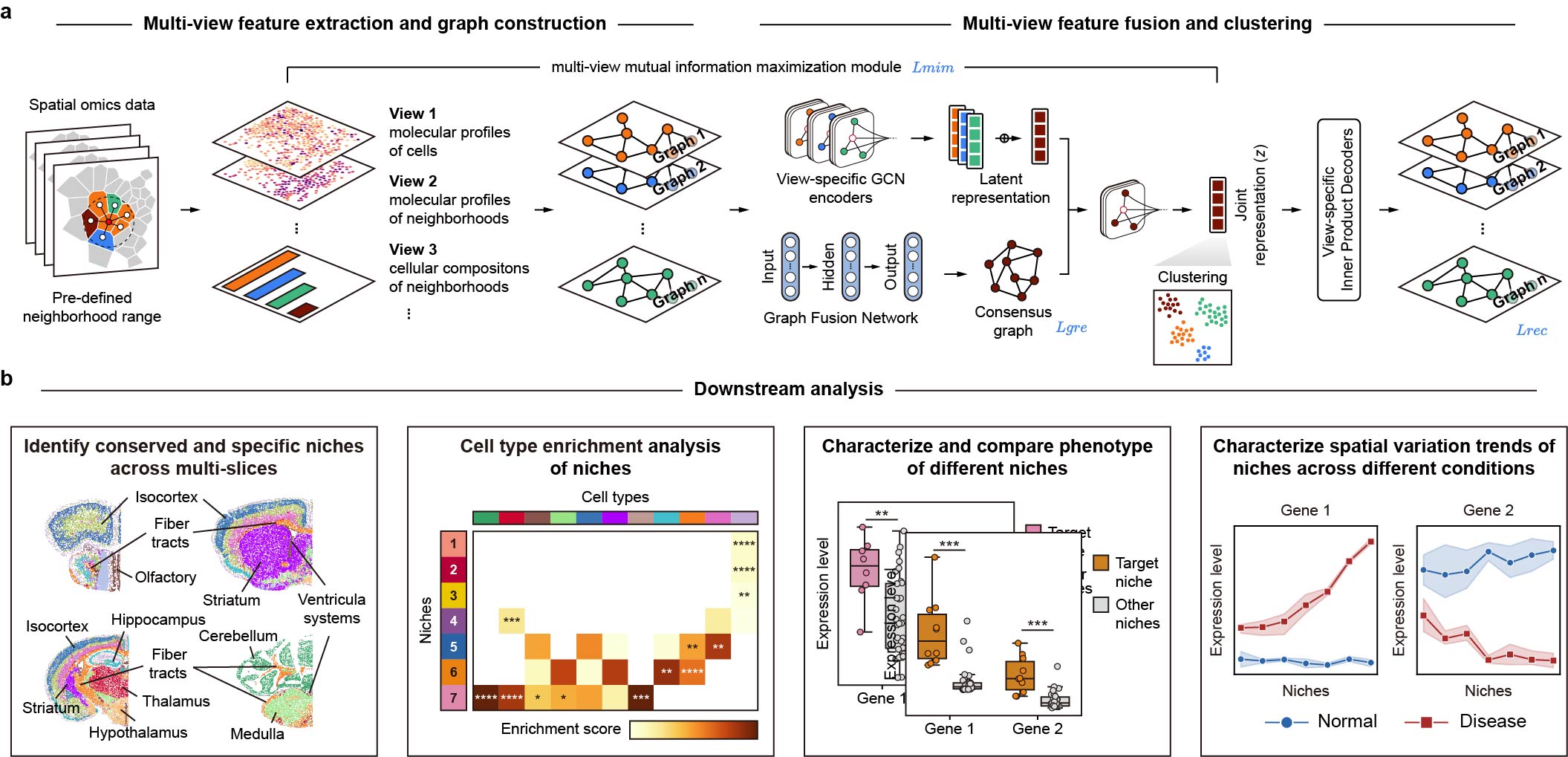
Identification and characterization of cell niches in tissue from spatial omics data at single-cell resolution
scNiche is a computational framework to identify and characterize cell niches from single-cell spatial omics data
For scNiche, the Python version need is over 3.9. If you have already installed a lower version of Python, consider installing Anaconda, and then you can create a new environment.
cd scNiche-main
conda env create -f scniche_dev.yaml -n scniche
conda activate scniche
We developed scNiche in a CUDA 11.3 environment. Here is an example of installing PyTorch and DGL with CUDA11.3:
# install PyTorch
pip install torch==1.12.1+cu113 --extra-index-url https://download.pytorch.org/whl/cu113
# install DGL
pip install dgl==1.1.0+cu113 -f https://data.dgl.ai/wheels/cu113/repo.html
The version of PyTorch and DGL should be suitable to the CUDA version of your machine. You can find the appropriate version on the PyTorch and DGL website.
python setup.py build
python setup.py install
By default, scNiche requires the single-cell spatial omics data (stored as .h5ad format) as input, where cell population label of each cell needs to be provided.
Here are examples of scNiche on simulated and biological datasets:
- Demonstration of scNiche on the simulated data
- Demonstration of scNiche on the mouse V1 neocortex STARmap data
scNiche also provides a subgraph-based batch training strategy to scale to large datasets and multi-slices:
- Batch training strategy of scNiche for single-slice:
- Demonstration of scNiche on the mouse spleen CODEX data (over 80,000 cells per slice)
- Batch training strategy of scNiche for multi-slices:
- Demonstration of scNiche on the human upper tract urothelial carcinoma (UTUC) IMC data (containing 115,060 cells from 16 slices)
- Demonstration of scNiche on the mouse frontal cortex and striatum MERFISH data (containing 376,107 cells from 31 slices)
We here take 4 slices from the same donor of the human DLPFC 10X Visium data as an example.
In contrast to spatial proteomics data, which usually contain only a few dozen proteins, these spatial transcriptomics data can often measure tens of thousands of genes, with potential batch effects commonly present across tissue slices from different samples. Therefore, dimensionality reduction and batch effect removal need to be performed on the molecular profiles of the cells and their neighborhoods before run scNiche. We used scVI by defalut, however, simple PCA dimensionality reduction or other deep learning-based integration methods like scArches are also applicable.
Furthermore, cell type labels are usually unavailable for these spatial transcriptomics data. As alternatives, we can:
- Use the
deconvolution results of spotsas a substitute view to replace thecellular compositions of neighborhoods. We used the human middle temporal gyrus (MTG) scRNA-seq data by Hodge et al. as the single-cell reference, and deconvoluted the spots using Cell2location:
- Only use the molecular profiles of cells and neighborhoods as input:
Multi-slice analysis of 4 slices based on the batch training strategy of scNiche:
The strategy of scNiche for modeling features from different views of the cell offers more possible avenues for expansion, such as application to spatial multi-omics data. We here ran scNiche on a postnatal day (P)22 mouse brain coronal section dataset generated by Zhang et al., which includes RNA-seq and CUT&Tag (acetylated histone H3 Lys27 (H3K27ac) histone modification) modalities. The dataset can be downloaded here.
scNiche also offers a downstream analytical framework for characterizing cell niches more comprehensively.
Here are examples of scNiche on two biological datasets:
- Demonstration of scNiche on the human triple-negative breast cancer (TNBC) MIBI-TOF data
- Demonstration of scNiche on the mouse liver Seq-Scope data
The scNiche model is developed based on the multi-view clustering framework (CMGEC). We thank the authors for releasing the codes.
scNiche is developed by Jingyang Qian. Should you have any questions, please contact Jingyang Qian at [email protected].
Qian, J., Shao, X., Bao, H. et al. Identification and characterization of cell niches in tissue from spatial omics data at single-cell resolution. Nat Commun 16, 1693 (2025). https://doi.org/10.1038/s41467-025-57029-9